Understanding viral assembly with quantitative super-resolution microscopy: from single-molecule imaging to fast live-cell imaging
- Abstract number
- 36
- Presentation Form
- Oral
- Corresponding Email
- [email protected]
- Session
- Super-Resolution Microscopy
- Authors
- Romain Laine (1, 2)
- Affiliations
-
1. MRC-LMCB, UCL
2. Francis Crick Institute
- Keywords
super-resolution, viral replication, virus assembly, quantitative analysis, single-particle averaging, machine learning, molecular counting
- Abstract text
Understanding viral assembly with quantitative super-resolution microscopy: from single-molecule imaging to fast live-cell imaging
Romain F. Laine 1,2, Anna Albecka-Moreau 3, Yuen Yuen 1, Caron Jacobs 1, Tommaso Galgani 4, Colin Crump 3, Clemens F. Kaminski 5, Bassam Hajj 4, Mark March 1, Ricardo Henriques 1,2,6
1 MRC-LMCB, UCL, London, UK
2 Francis Crick Institute, London, UK
3 Division of Virology, Department of Pathology, Cambridge University, Tennis Court Road, Cambridge, CB2 1QP, UK
4 Laboratoire Physico-Chimie, Institut Curie, CNRS UMR168, Paris, France
5 Department of Chemical Engineering and Biotechnology, Laser Analytics Group, Cambridge University, Pembroke Street, Cambridge, CB2 3RA, UK
6 Instituto Gulbenkian de Ciência, 2780-156 Oeiras, Portugal
Summary
Due to their small sizes, the study of viral assembly has historically been carried out using electron microscopy. In the past decades, super-resolution technologies have allowed the imaging of viral replication with fluorescence microscopy while maintaining high molecular specificity and live-cell compatibility. Here, we show that nanoscale imaging can be combined with bespoke quantitative analysis to unravel detailed viral assembly observations at the molecular level. In particular, we show applications of single-particle averaging, machine learning and molecular counting as powerful tools for peering into the molecular life of viral replication. We finally show an outlook at how the development of fast live-cell super-resolution may also contribute to observations of dynamics nanoscale events during infection.
Introduction
Viruses are obligate pathogens that commonly infect eukaryotic cells or bacteria. They typically have sub-micron sizes that make them difficult to image with standard light microscopy. The method of choice to study the structure of viral components has historically been electron microscopy. This approach, however, has the downside of having relatively poor molecular specificity and no live-cell compatibility. This, therefore, limits what can be observed during viral replication.
In the past decades, the development of super-resolution microscopy (SRM) has uniquely allowed observations of viral components at the nanoscale 1,2. It has shown great potential for viral replication imaging in relevant biological contexts. Beyond achieving nanoscale resolution, SRM can be combined with quantitative approaches and provide a deep understanding of biological systems. Here, we show that combining SRM with single-particle averaging 3, machine learning 4 and molecular counting 5 can provide ultra-structural information about viral assembly, large scale understanding of viral population and insights into receptor reorganisation upon infection.
Materials and Methods
Here, we combine SRM methodologies, such as single-molecule localization microscopy (SMLM) and Structured Illumination Microscopy (SIM), to decipher nanoscale structural features involved in viral replication. Image and data analysis tools based on single-particle averaging, machine learning and molecular counting allow the extraction of quantitative understanding of the systems.
Results
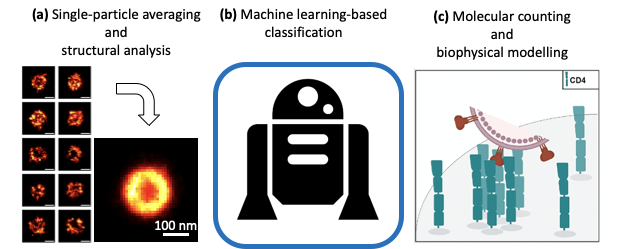
First, by combining SMLM with single-particle average and modelling, we show such data can extract the position of protein layers with near-Angstrom resolution within the tegument layer of Herpes Simplex Virus type 1 (HSV-1) 3. Second, we show that fast and high-throughput SIM imaging combined with machine-learning-based classification can describe the structural features of heterogeneous viral populations such as influenza and Newcastle Disease Virus (NDV) 4. This has a characterisation application in the field of biomedical industry. Last, we exploit the quantitative single-molecule imaging capabilities of SMLM to observe and quantify the nanoscale re-organisation of viral receptors at T cells’ surface upon HIV-1 binding 5.
Figure 1: Quantitative analyses applied to virus replication study. (a) Single-particle averaging, (b) machine learning and (c) molecular counting.
Conclusion and outlook
Taken together, we show that SRM and quantitative analysis have great potential in understanding viral replication at the nanoscale. Additionally, we propose that refinement of live-cell compatible SRM methods like SRRF and in particular a novel implementation of it allowing 3D imaging may offer in the future an avenue for studying important stages of infection at the nanoscale within infected cells, therefore giving access to much-sought-after dynamics.
References
- References