Guided- Deconvolution in Correlative Light and Electron Microscopy
- Abstract number
- 41
- Presentation Form
- Oral
- DOI
- 10.22443/rms.elmi2021.41
- Corresponding Email
- [email protected]
- Session
- Correlative Microscopy Across the Scales
- Authors
- Fengjiao Ma (2, 5, 4), Sedzicki Jaroslaw (1), Stephanie Hoeppener (3, 4), Christoph Dehio (1), Rainer Heintzmann (2, 5, 4)
- Affiliations
-
1. Biozentrum, University of Basel, 4056 Basel, Switzerland
2. Institute of Physical Chemistry and Abbe Center of Photonics, Friedrich Schiller University Jena, Jena, Germany
3. Organic and Macromolecular Chemistry (IOMC), Friedrich Schiller University Jena, Jena, Germany
4. Jena Center for Soft Matter (JCSM), Friedrich Schiller University Jena, Jena, Germany
5. Leibniz Institute of Photonic Technology, Jena, Germany
- Keywords
correlative light and electron microscopy, deconvolution
- Abstract text
This project is aimed at investigating optimized approaches to obtain molecule specific information by the (fluorescence) light microscopy (LM) measurement and to join it with the ultra-structure seen in its correlated the electron microscopy (EM) images.
Correlative light and electron microscopy is popular for investigating the internal structure of cells[1,2] By specific labeling of the proteins of interest, one can easily study their role in the cell. However, even with current super resolution microscope technology, ultra-structural detail in the cell is still difficult to discern in light microscopy. The use of the electron microscopy enables to further observe the cell's microstructure down to the nanoscale level. However, transmission electron microscopy's low and relatively unspecific contrast makes it difficult to conclude on the role of individual molecules.
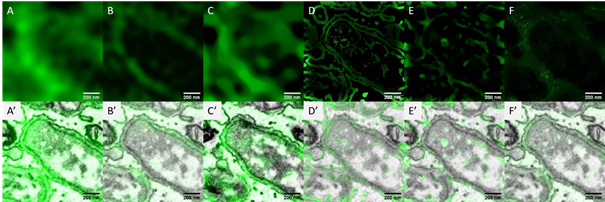
We implement a customized algorithm to enable an EM-guided deconvolution, which can automatically recognize the LM labeled structures in and EM image. This will, for example, help the study of changes in the ultrastructural environment in response to nanoparticle uptake. We demonstrate that our EM-guided deconvolution of light microscopy images, achieves better results than state of the art regularized deconvolution [3] of LM data only (Fig.1). Both, simulated and experimental results will be presented.
Figure 1: A comparison of the measured light microscopy image and the deconvolution reconstruction results. The image shows the GFP labeled endoplasmic reticulum (ER) when the facultative intracellular pathogen Brucella enter into host cells [4]. A) measured wide field fluorescence light microscopy image; B) structure illumination microscopy[5] image; C)-F) the restored images of (A) using the modified total variation deconvolution, the entropy-based intensity-guided deconvolution, the modified total variation deconvolution combining intensity-guided deconvolution and the gradient-guided deconvolution. A’) – F’) the overlay of the measured / restored images and the electron microscopy image. The restored imaged from the EM-guided deconvolution (D-F) represent more details about the ER membranes than both the regularized deconvolution (C) and the super resolution light microscopy image (B).
- References
[1] Jahn, K. A., et al. "Correlative microscopy: providing new understanding in the biomedical and plant sciences." Micron 43.5 (2012), 565-582.
[2] Ando, Toshio, et al. "The 2018 correlative microscopy techniques roadmap." Journal of physics D: Applied physics 51.44 (2018), 443001.
[3] Soulez, Ferréol, et al. "Blind deconvolution of 3D data in wide field fluorescence microscopy." 2012 9th IEEE International Symposium on Biomedical Imaging (ISBI). IEEE, 2012.
[4] Sedzicki, Jaroslaw, et al. "3D correlative electron microscopy reveals continuity of Brucella-containing vacuoles with the endoplasmic reticulum." Journal of cell science 131.4 (2018).
[5] Jost, Aurélie, and Rainer Heintzmann. "Superresolution multidimensional imaging with structured illumination microscopy." Annual Review of Materials Research 43 (2013), 261-282.